SyNaToSe
Leveraging Cross-Domain Synergies for Efficient Machine Learning of Nanoscale Tomogram Segmentation
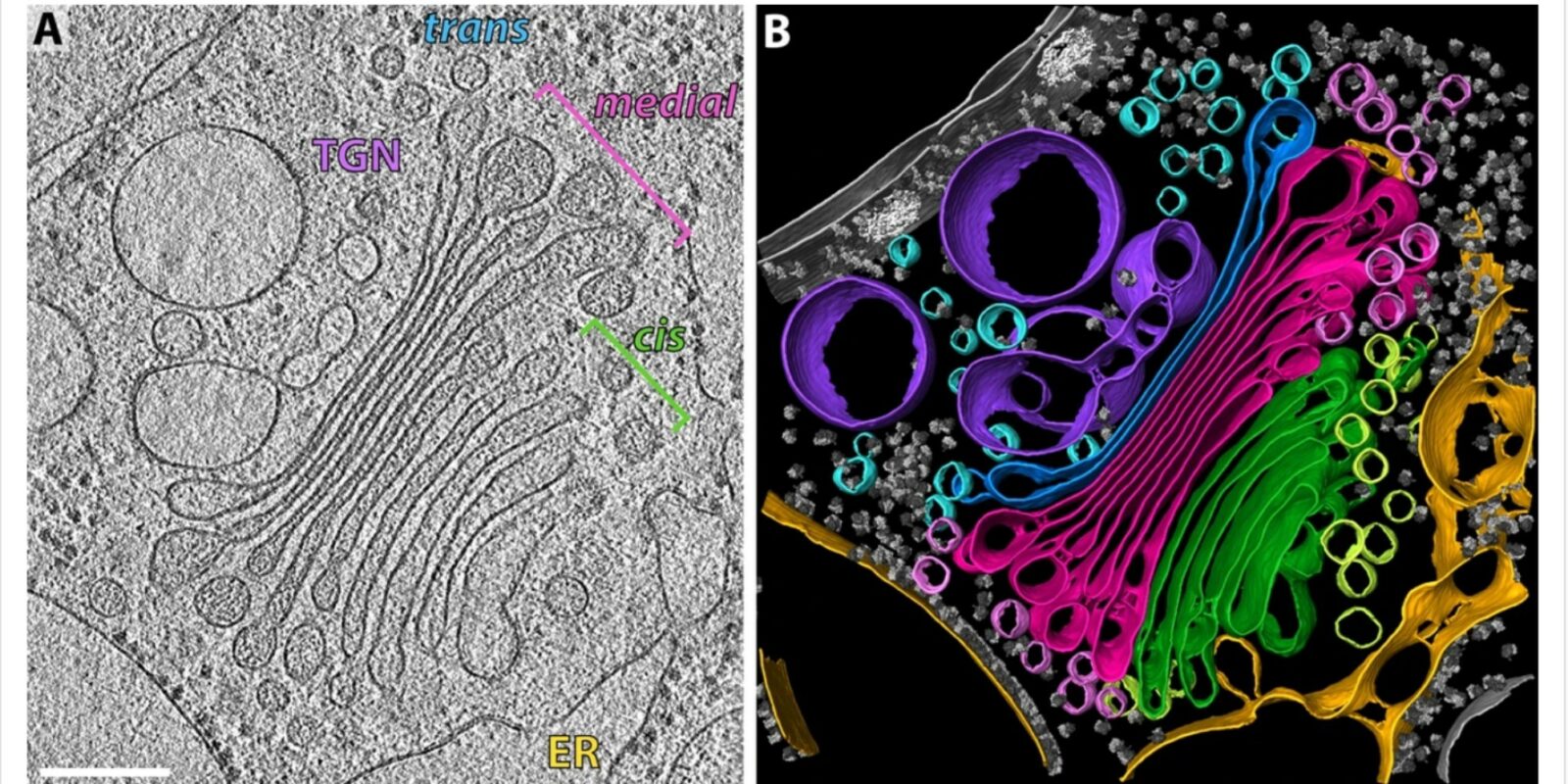
Before you can get an artificial intelligence to detect certain things automatically, you have to teach it first. For example, if you want a computer to learn how to recognise specific structures inside cells, you first need to feed it with a few dozen images of cells taken under a microscope, in which you have marked, or annotated, those structures by hand. “But, creating these annotations for the computer is often incredibly laborious,” says Dr. Dagmar Kainmüller of the Max Delbrück Center for Molecular Medicine. Therefore, she and her colleagues are working on a method for teaching a machine using little or no annotated data at all.
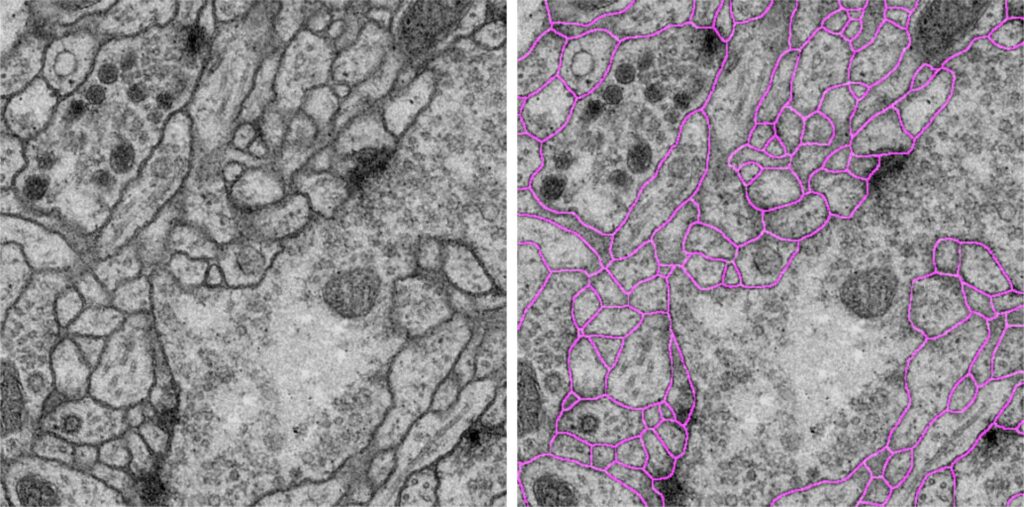
The team is currently working from images of cells taken at nanometre resolution under a cryo-electron microscope. Researchers use these images to identify specific membrane-like cell structures – and can give the job of finding them to an artificial intelligence. “There are several groups at a number of Helmholtz institutes working with images from cryo-electron microscopes,” Dagmar Kainmüller says.
Yet, so far, they have to spend weeks or even months preparing these images before they can be used to train the artificial intelligence for each new structure. The aim is therefore to train an algorithm so that it can be used for this and other related search tasks as well – in other words, for capturing similar structures from different imaging techniques or for capturing different structures from similar imaging techniques.
Other projects

HIT Permafrost
The Hidden Image of Thawing Permafrost
The project aims to develop a method for determining just how extensively thaw processes have already progressed in permafrost regions. The machine learning approach to be developed will be used to analyse radar images from aircraft in order to learn more about the properties of the subsurface permafrost.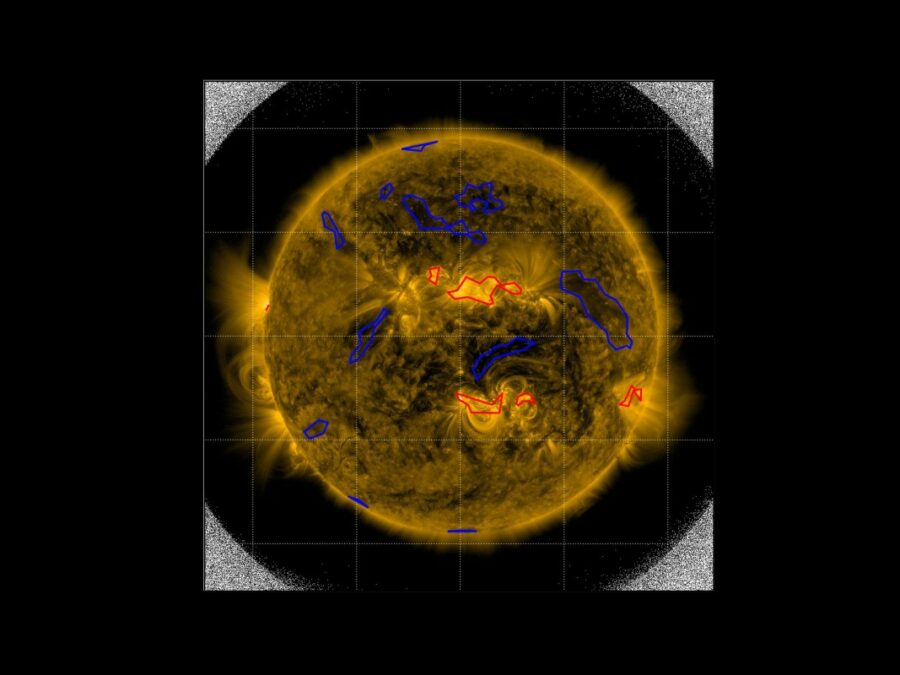
SIM
Solar Image-based Modelling
The aim of the project is to develop an algorithm by which computers can automatically predict the space weather. This will make use of datasets of solar images that have been captured from space. The method could replace computationally demanding physics-based models and deliver space weather forecasts long before the effects of solar events are […]
SATOMI
Tackling the segmentation and tracking challenges of growing colonies and microbialdiversity
An artificial intelligence will observe the growth of bacteria: from microscope images of bacterial cultures taken at regular intervals, it will precisely track the development and division of individual cells – even when multiple bacterial species are cultivated together.