EMSIG
Event-driven Microscopy for Smart Microfluidic Single-cell Analysis
Microfluidic live-cell imaging (MLCI) unlocks spatio-temporal insights into population heterogeneity emerging from a single cell. Deep-learning powered image analysis, developed in the HI project SATOMI, facilitates offline analysis of high-quality data. For becoming a versatile screening tool, MLCI must master the challenge of robust real-time analysis, capable to detect rare events in hundreds of parallel experiments and resolve their temporal evolution.
Towards this, EMSIG brings smart live-event detection capabilities to MLCI to facilitate the adaptive optimization of biological event resolution and autonomously counteracting deteriorating image qualities, as examples. The distinct high-risk factor is in the robust and accurate real-time classification of events at the interface between imaging, image analysis and biology. If successful, to date unavailable insights into biological processes are unlocked and the technology readiness level of microfluidic live-cell analysis boosted.
Other projects
SATOMI
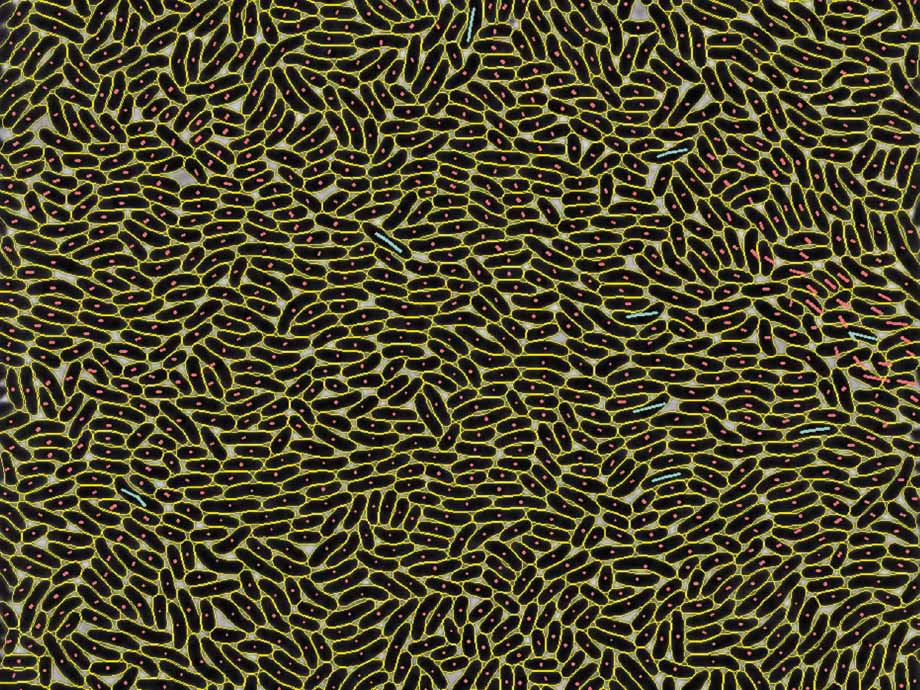
Tackling the segmentation and tracking challenges of growing colonies and microbialdiversity
An artificial intelligence will observe the growth of bacteria: from microscope images of bacterial cultures taken at regular intervals, it will precisely track the development and division of individual cells – even when multiple bacterial species are cultivated together.BENIGN
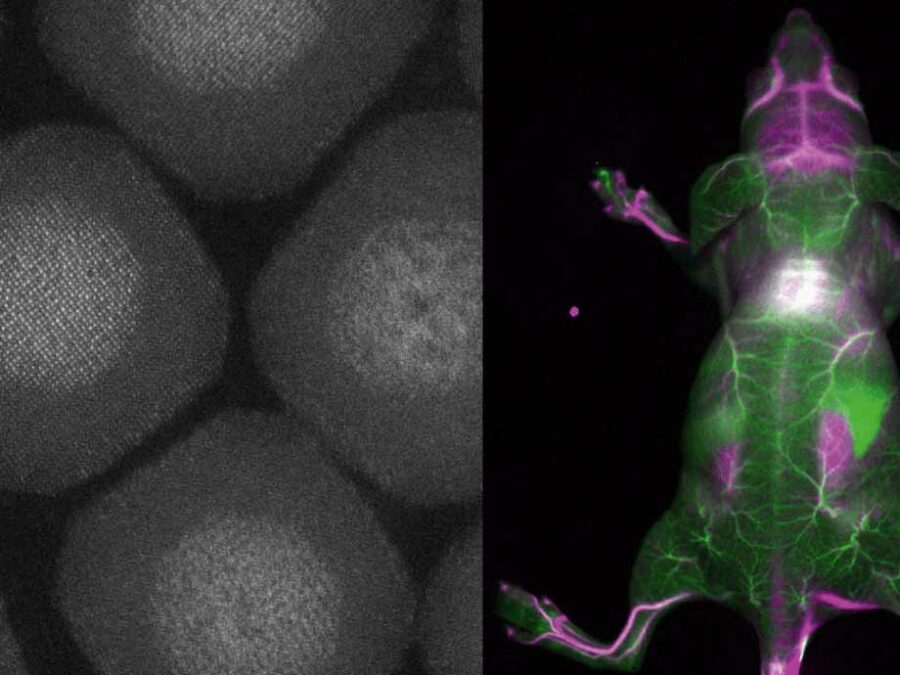
Biocompatible and Efficient Nanocrystals for Shortwave Infrared Imaging
The BENIGN project aims to enable non-invasive molecular imaging with cellular resolution in vivo at depths of several millimeters. This will be achieved using light from the shortwave infrared (SWIR) range (1000-2000 nm), which has less scattering and autofluorescence compared to the visible and near-infrared spectral range. Bright and targeted imaging agents are needed to fully exploit this range. The project will develop a new approach using lanthanide-based core-shell structures that emit light in the 1500-2000 nm range.SIM
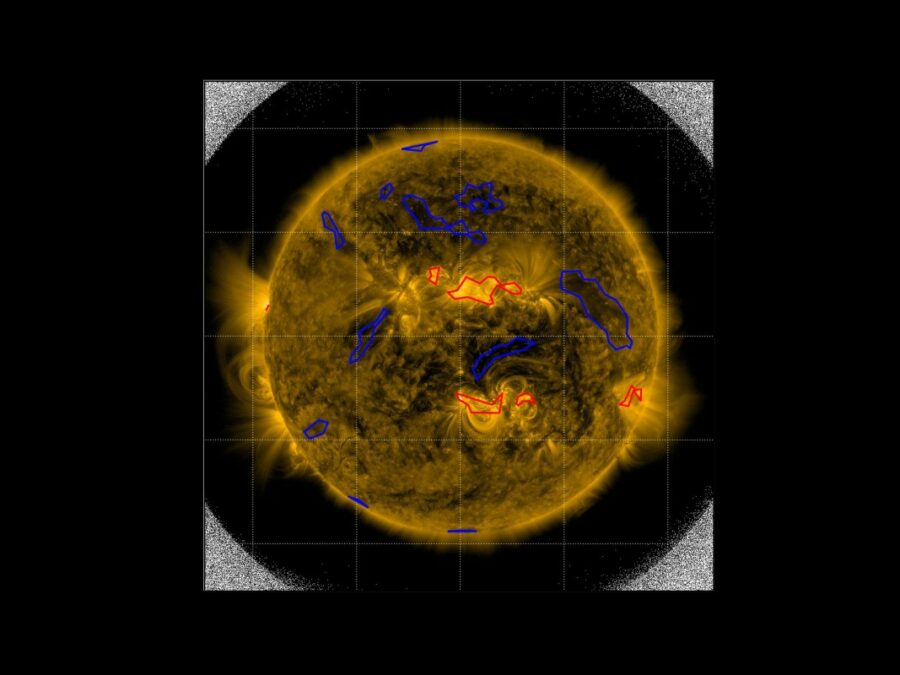
Solar Image-based Modelling
The aim of the project is to develop an algorithm by which computers can automatically predict the space weather. This will make use of datasets of solar images that have been captured from space. The method could replace computationally demanding physics-based models and deliver space weather forecasts long before the effects of solar events are […]