SCHEMA – profiling Spatial Cancer HEterogeneity across modalities to benchmark Metastasis risk prediction
What is the project about?
Early detection of cancer metastases is critical, yet robust predictive and actionable occurrence biomarkers are still lacking. We will build the largest single-cell, spatial, multimodal benchmarking resource for metastasis prediction, aggregating public datasets of primary and metastatic lung, colon and breast tumors with newly generated data.
What main scientific or societal challenge does the benchmark address?
Metastases represent a significant exacerbation of tumor severity. If one could predict the likelihood of tumors metastasizing, this could inform treatment decisions to avoid or delay this outcome. SCHEMA develops a benchmark dataset of primary tumor samples and metadata on whether the tumor has metastasized at different time points after sampling. With this dataset, a challenge for machine learning scientists will be defined to build prognostic models for likelihood of tumors metastasizing, promoting innovation in prognostic modeling for a clinically relevant task.
What gap in the scientific community led to the creation or expansion of this benchmarking dataset?
Metastasis in cancer is one of the main determinants of poor outcome. They can arise very early in the process of cancer progression and yet often remain undetected at diagnosis. Knowing which cancer patient is more likely to develop metastases will enable targeted early screening for improved diagnostics and the adaptation of treatment regimes in a personalized manner. Spatial niches can serve as potential biomarkers for cancer outcome or risk of metastases, however harmonized datasets of sufficiently large cohorts that can be used for training machine learning models to evaluate these prediction tasks are lacking. The SCHEMA benchmarking dataset of primary and metastatic tumors will provide this resource, focusing on three highly prevalent cancers with high metastasis risk – lung, colon and breast.
How does the benchmark dataset support reproducibility, robustness, and fairness in AI research?
Providing access to key data- and benchmark datasets for the community will help us not only to disseminate our findings broader and faster, but will also empower researchers of multiple disciplines to bring their expertise to bear to tackle this clinical challenge.
What is the project’s structure — from data curation to expected outputs such as publications or competitions?
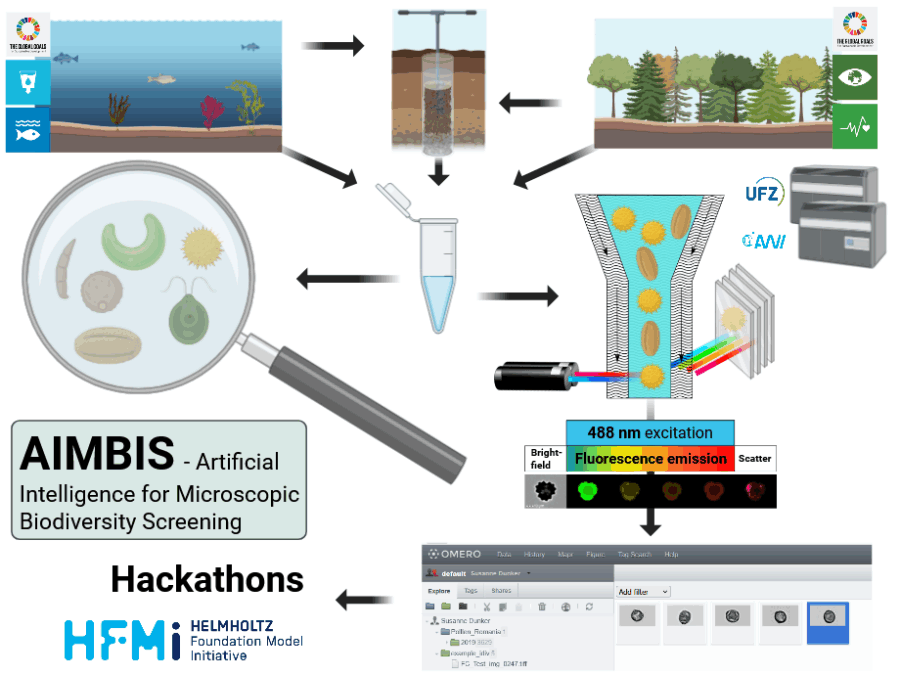
The SCHEMA benchmark dataset generation will be performed in two stages: first we collect public datasets of cancer spatial transcriptomics and proteomics and secondly, we will perform rapid, extensive data generation effort that fills the gaps in available datasets and metadata completeness. For this, a set of cancer samples will be profiled with spatial transcriptomics and proteomics on consecutive slides with single-cell resolution. Finally, we will jointly process these datasets and harmonize the metadata, upload them to an industry-grade spatial omics database, and organize a community hackathon to promote the use of the SCHEMA dataset by the scientific community on the OpenProblems in Single-Cell Analysis platform. The hackathon will promote and encourage a wider community to use the dataset, and we will prepare a manuscript with the benchmarking data, the living benchmark, and the hackathon results.
In what ways does the project foster cross-domain, cross-center, or interdisciplinary collaboration?
Given the complexity of the disease and the clinical condition we are interested in, one realises that an isolated laboratory will not be able to provide groundbreaking insights. The close interaction of multiple cross- and interdisciplinary centers and groups, however, has the potential to unlock new avenues, hopefully towards the identification of not only predictive markers but also actionable targets.
To this end have curated a team, encompassing AI specialists, molecular biologists, clinicians, toxicologists and geneticists, located throughout Germany and spanning multiple Helmholtz Centers and additional partner sites.
Making our data and findings public and easily accessible will empower and allow researchers not only of the fields mentioned adobe but e.g. from the Environmental Health Cluster, to incorporate our findings into their research projects and cross-reference with the existing patient cohorts readily available, allowing for epidemiological analysis and in-depth risk assessment.
What impact does the project aim to achieve — within Helmholtz and across the broader research and industry community?
We envision that our project will shed new light on the pathophysiological mechanism of solid tumor metastasis, a clinical challenge with currently limited biomarker availability. The possibility to identify new predictive markers would allow for an acceleration of patient stratification and could be used for an informed adaptation of treatment regimens. Furthermore, we do hope to identify actionable targets.
Not only could this spark new research topics within the realm of metastatic research, but would bear fruit with regard to initiation of drug development and small molecule design via the available world leading units available at Helmholtz. Furthermore, and in the long run, do we envision to collaborate with industry to bring our findings to the clinics, full circle from the hospital bed to the bench and back to the bedside.
Other projects
BASE: Benchmarking Agro-environmental database for Sustainable agriculture intensification
Building a BASE dataset enables robust predictions of yield potential, resource efficiency, and sustainability thresholds, driving climate resilience and sustainable agricultural intensification