AMOEBE: lArge-scale Multi-mOdal Microbial livE-cell imaging BEnchmark
What is the project about?
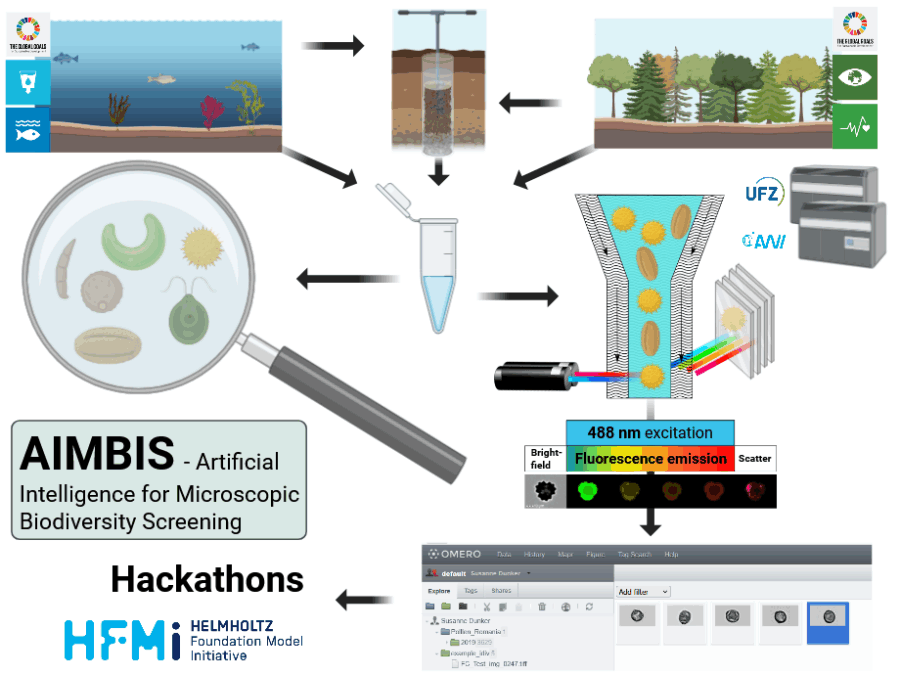
AMOEBE establishes a FAIR benchmark for AI-driven microbial imaging using high-resolution time-lapse microscopy. It combines unlabeled data for foundation models with high-quality expert-labeled subsets for tasks like cell segmentation and tracking, fostering benchmarking and cross-domain collaboration.
What motivated you to apply for UNLOCK, and how does the project align with the initiative’s vision?
We were motivated to apply for the hyperUNLOCK initiative because its vision of building cross-domain, FAIR, and AI-ready benchmark datasets directly aligns with our long-term goal of advancing data-driven microbiology and biotechnology research. With AMOEBE, we aim to unlock the potential of AI in microbial live-cell imaging by establishing a standardized, open benchmark dataset that integrates high-quality microscopy data, expert annotations, and reproducible ML workflows. This contributes to hyperUNLOCK’s mission of fostering benchmarking science, interoperability, and collaboration across Helmholtz centers, enabling the broader life science community to leverage AI for making discoveries faster.
What gap in the scientific community led to the creation or expansion of this benchmarking dataset?
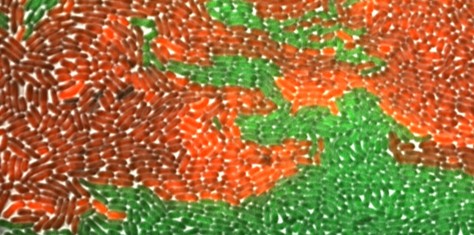
Microbial imaging lacks large, standardized benchmarks that capture the complexity of microbial populations. Existing datasets focus mainly on mammalian cells, overlooking challenges such as dense colony growth, frequent divisions, and low contrast. AMOEBE aims to bridge this gap by providing a high-quality benchmark to eventually advance and evaluate AI methods in this field.
How does the benchmark dataset support reproducibility, robustness, and fairness in AI research?
AMOEBE promotes reproducibility and robustness through open access and standardized metadata. Expert-validated annotations and diverse microbial data ensure fair, generalizable AI model evaluation across species, imaging modalities, and experimental setups.
What is the project’s structure — from data curation to expected outputs such as publications or competitions?
AMOEBE follows a structured, five-step workflow.
- Data collection and curation: We compile large-scale time-lapse microscopy datasets from diverse microbial species, cultivation systems, and imaging modalities, ensuring standardized metadata and quality control.
- Annotation: Expert-guided, semi-automated annotation generates high-quality ground truth for cell segmentation and tracking tasks.
- Integration: Data and labels are integrated into AI-ready formats (e.g., PyTorch loaders) with standardized train/validation/test splits.
- Benchmarking: Baseline implementations and biologically meaningful evaluation metrics are developed for reproducible performance assessment.
- Dissemination: The benchmark will be openly published under FAIR principles and promoted through community channels to invite hackathons or challenges to stimulate method comparison.
In what ways does the project foster cross-domain, cross-center, or interdisciplinary collaboration?
AMOEBE unites microbiology, imaging, AI, and data management experts from multiple Helmholtz centers and academia. Its FAIR, open design fosters cross-domain collaboration by combining biological and computational expertise and enabling shared use and expansion of the dataset across disciplines and institutions.
What impact does the project aim to achieve — within Helmholtz and across the broader research and industry community?
AMOEBE will establish a new standard for AI-driven microbial imaging by providing the first large-scale benchmark dataset of microbial time-lapse microscopy. Within Helmholtz, it will strengthen collaboration between life science and AI research, serving as a model for data-centric initiatives. Beyond Helmholtz, AMOEBE will enable researchers to develop, compare, and validate algorithms on realistic microbial data, accelerating innovation in biotechnology, health, and environmental sciences.