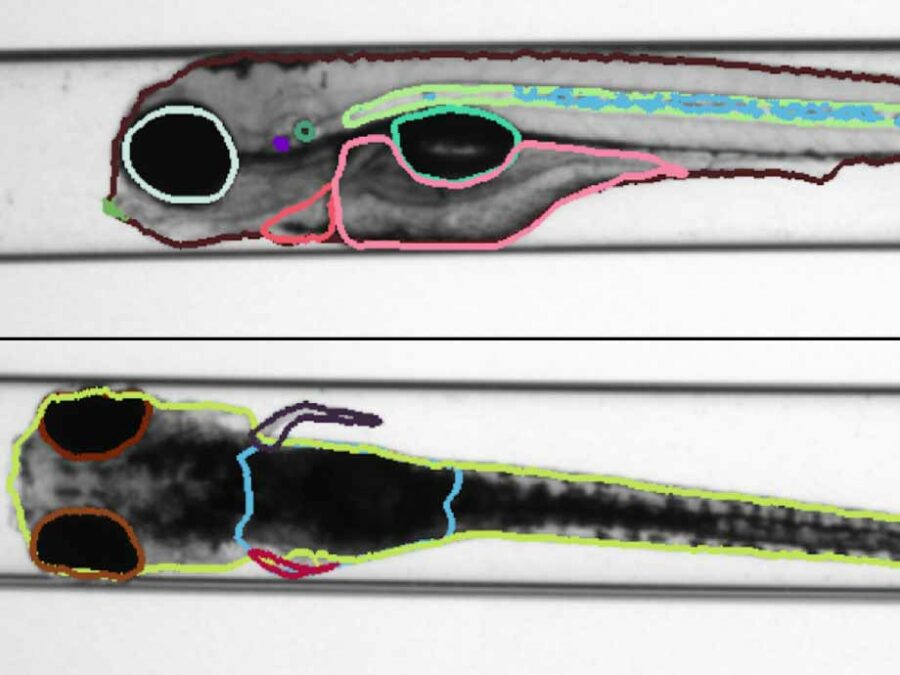
Polarity-JaM
Cell polarity involves the asymmetric distribution of cellular components, cell shape, and contacts with neighboring cells. Gradients and mechanical forces bias cell polarity, coordinated by communication between adjacent cells.
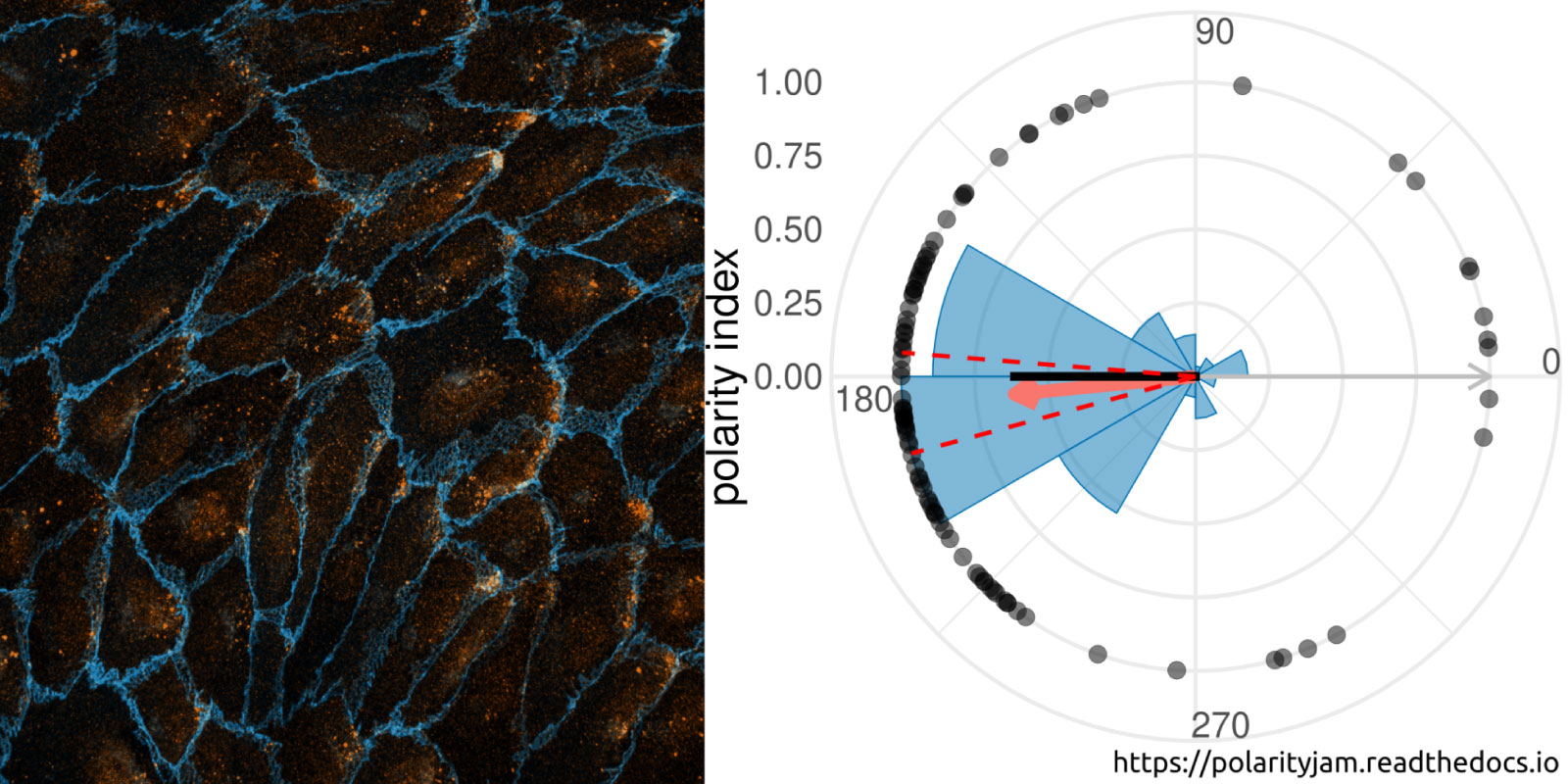
Advances in fluorescence microscopy and deep learning for image segmentation have expanded our understanding of cell polarity in health and disease. Polarity-JaM is an open-source package for exploratory image analysis. It offers a user-friendly interface for multi-channel single cell segmentation with deep learning algorithms, automatically extracting features like cell orientation and signaling molecule gradients. The web application computes circular statistics of cell polarity, such as polarity indices and circular correlation analysis, with comprehensive data visualization.
The focus is on fluorescence image data from endothelial cells (ECs) and their polarization behavior, relevant to vessel formation, repair, and diseases like cardiovascular conditions, cancer, and inflammation. The software’s architecture also supports other cell types and image modalities. Built in Python, Polarity-JaM integrates seamlessly into existing workflows, with a web application for statistical analysis and a Napari plugin available for exploratory analysis. More details can be found at https://polarityjam.readthedocs.io and www.polarityjam.com.
Other Collaborations
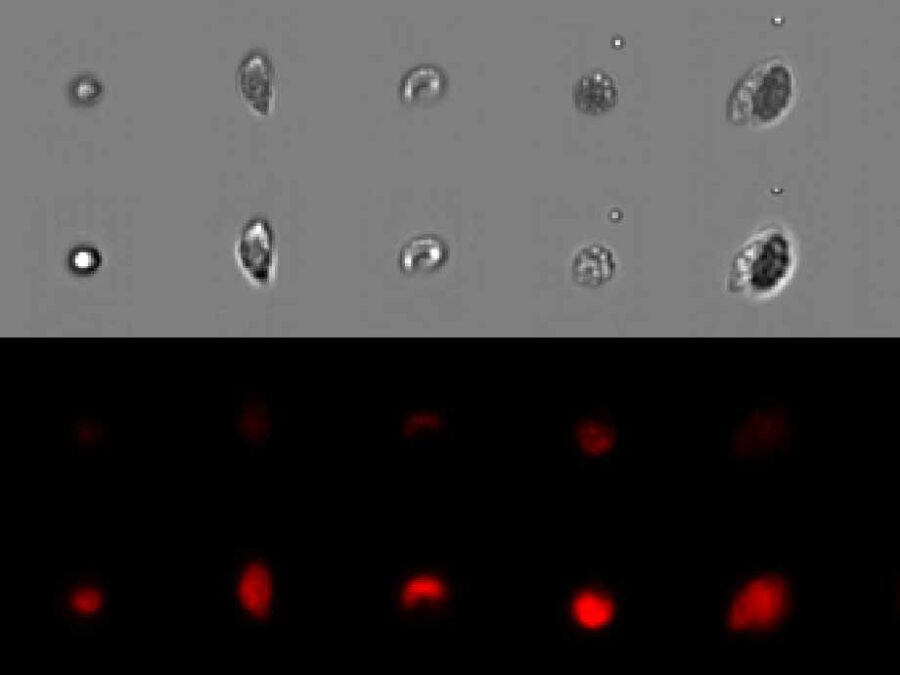
Automated Analysis of Evolutionary Experiments of Phytoplankton
There is a strong interest in understanding community assembly and dynamics. Experimental approaches using phytoplankton have proven to be extremely insightful to unravel underlying biological processes. Imaging flow cytometry is an emerging method becoming more and more popular in different fields. It allows us to capture changes within a community or population in a more […]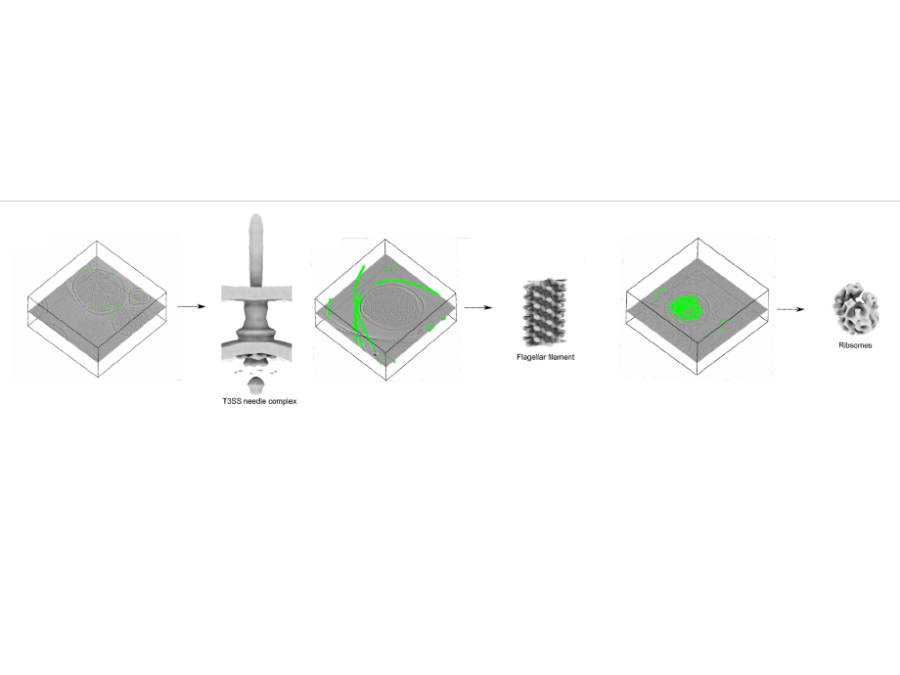
Pick Yolo
In collaboration with the Center for Structural Systems biology, Helmholtz Imaging has developed and trained a convolutional neural network for the picking of instances of proteins in cryoelectrontomograms (CryoET). The so picked instances are subsequently used to reconstruct a 3D structure of the proteins using subtomogram averaging methods. The novel picking methods exploit the well […]