Plaque Assays
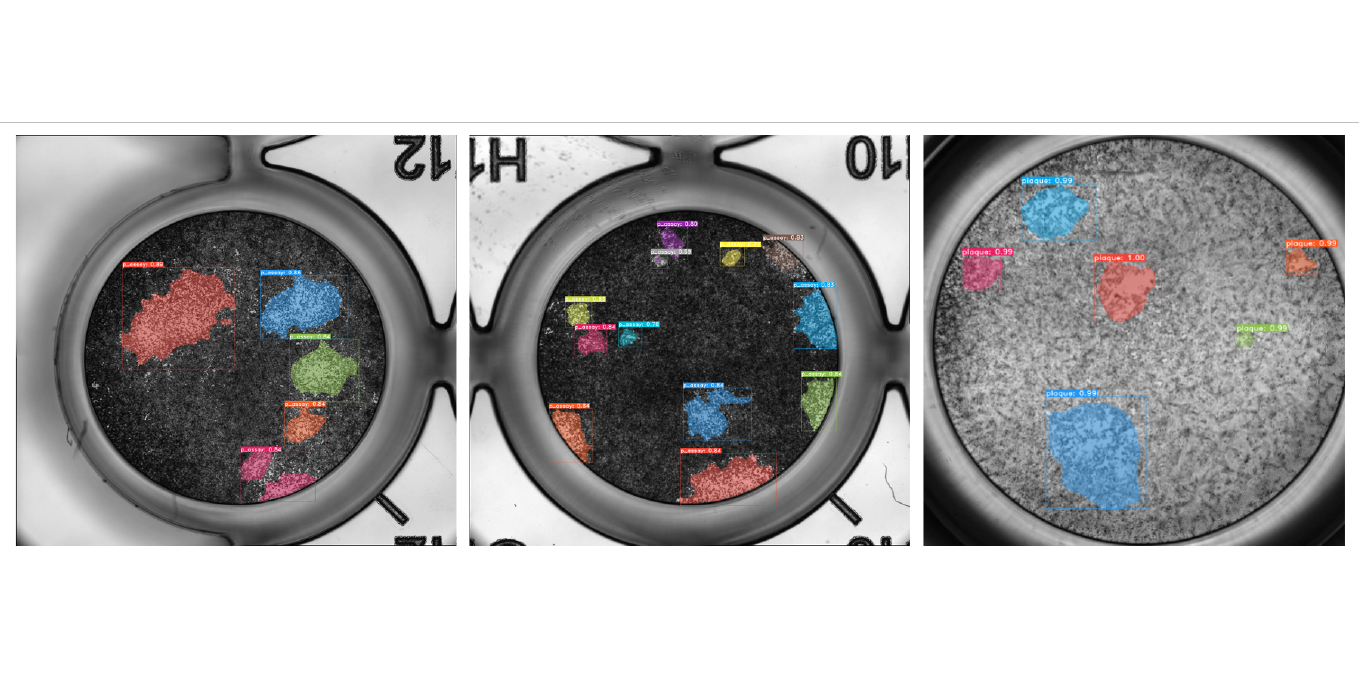
In collaboration with the Leibniz Institute of Virology and the Center for Structural Systems biology, Helmholtz Imaging is working on a processing pipeline for plaque assays. Plaque Assays are used to e.g. estimate the virus concentration in a given sample. For this high resolution images of cell cultures which have been infected with a virus containing solution, have to be analysed, plaques (i.e. areas of dead cells) need to be identified (instance segmentation) and counted. The challenge in this collaboration is the aim to provide a broadly applicable solution, capable to deal with various cell cultures, and viruses.
Other Collaborations
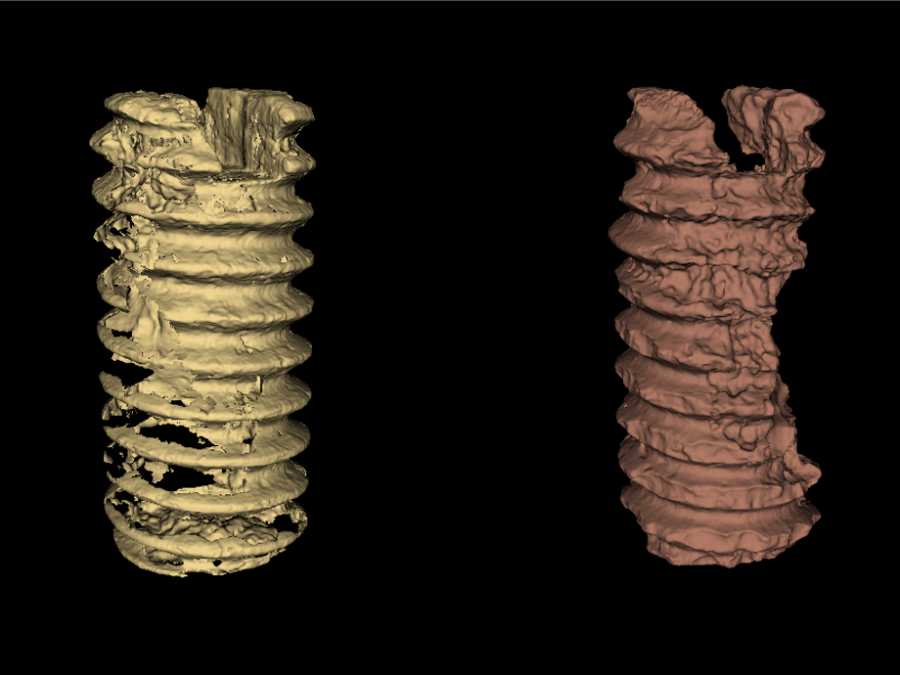
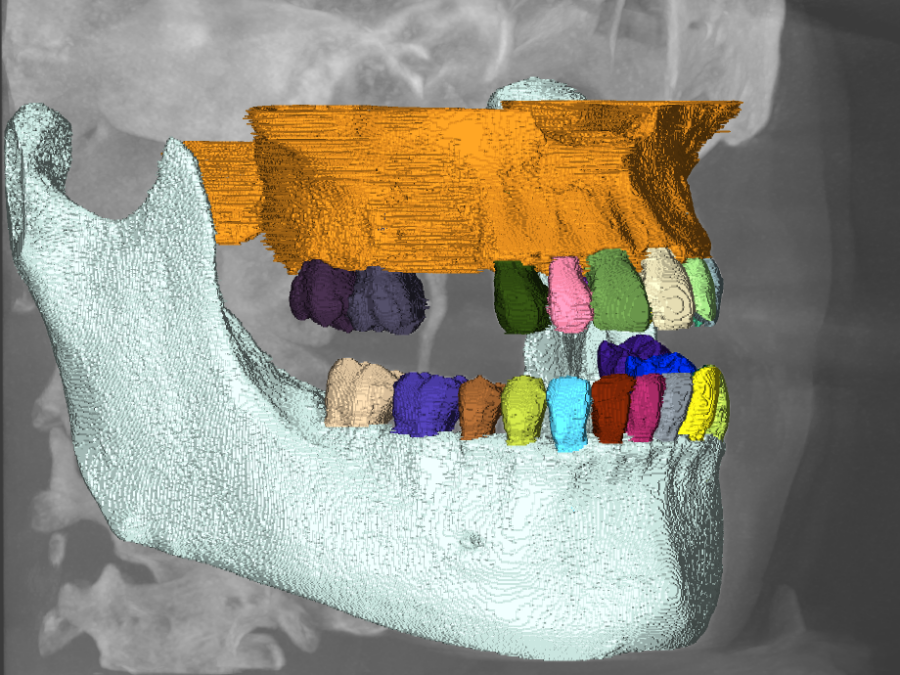
Segmentation of biodegradable bone implants
Together with the experts from Hereon the Helmholtz Imaging Support Team collaborates on segmenting synchrotron CT data. In 2021 the collaboration on semantic segmentation of biodegradable bone implants using a U-net resulted in two publications acknowledging the contribution of Helmholtz Imaging [1],[2]. Beyond that, we studied “Instance segmentation of paper fibers” imaged at the Hereon […]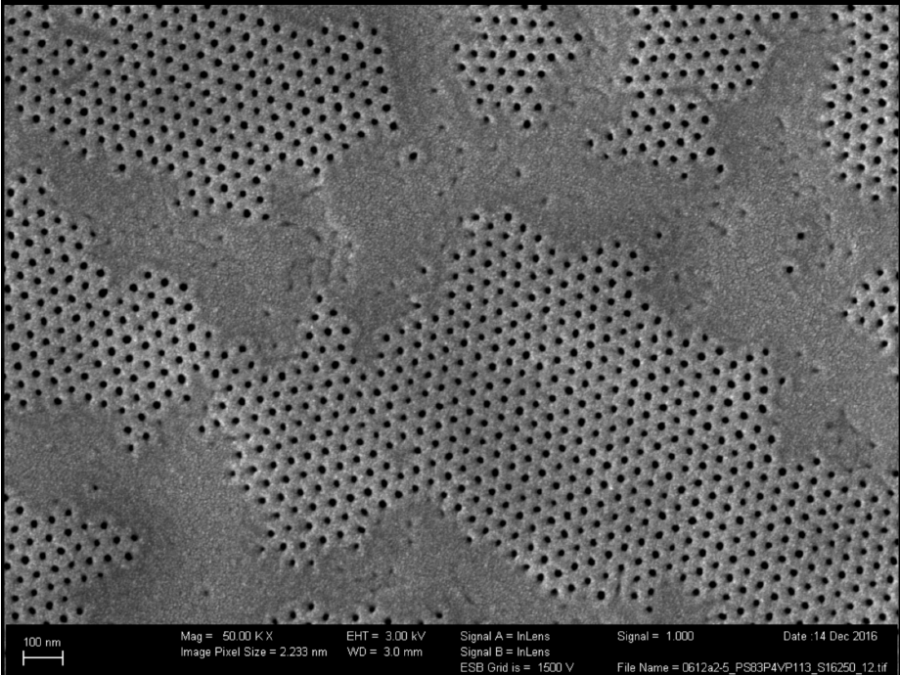
Connecting membrane pores and production parameters via machine learning (COMPUTING)
Isoporous block-copolymer membranes play a fundamental role in the filtration of liquids and can, for example, be used to purify drinking water. Despite recent progress in understanding the membrane formation process, finding suitable production parameters for a given precursor material (such as polymers of a certain length) still occurs in a trial-and-error fashion, wasting materials, […]