BetaSeg
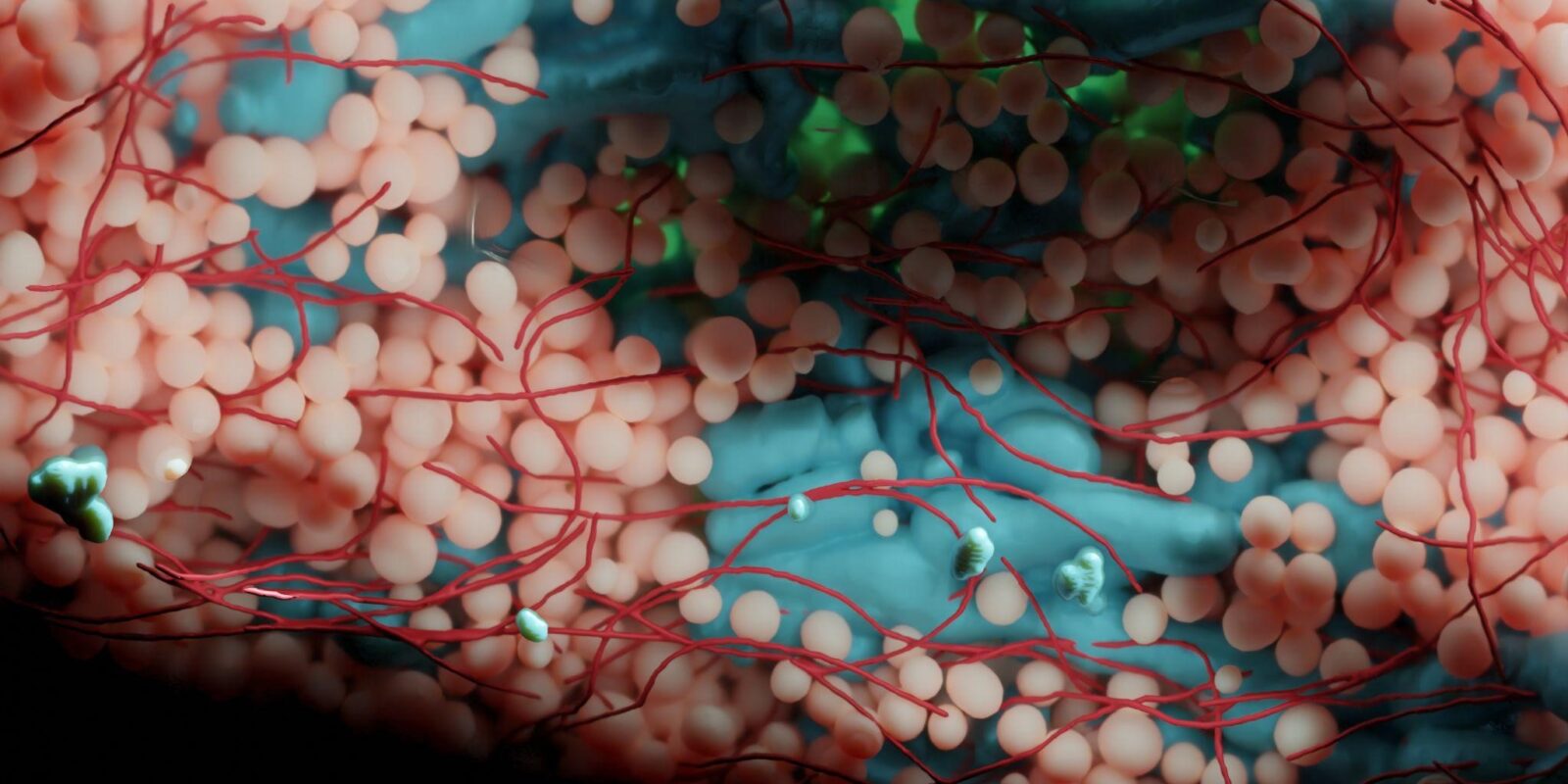
Volume electron microscopy is the method of choice for the in situ interrogation of cellular ultrastructure at the nanometer scale, and with the increase in large raw image datasets generated, improving computational strategies for image segmentation and spatial analysis is necessary. Here we describe a practical and annotation-efficient pipeline for organelle-specific segmentation, spatial analysis and visualization of large volume electron microscopy datasets using freely available, user-friendly software tools that can be run on a single standard workstation.
The procedures are aimed at researchers in the life sciences with modest computational expertise, who use volume electron microscopy and need to generate three-dimensional (3D) segmentation labels for different types of cell organelles while minimizing manual annotation efforts, to analyze the spatial interactions between organelle instances and to visualize the 3D segmentation results.
We provide detailed guidelines for choosing well-suited segmentation tools for specific cell organelles, and to bridge compatibility issues between freely available open-source tools, we distribute the critical steps as easily installable Album solutions for deep learning segmentation, spatial analysis and 3D rendering. Our detailed description can serve as a reference for similar projects requiring particular strategies for single- or multiple-organelle analysis, which can be achieved with computational resources commonly available to single-user setups.
Publication
Modular segmentation, spatial analysis and visualization of volume electron microscopy datasets by Andreas Müller et at.
Other Collaborations
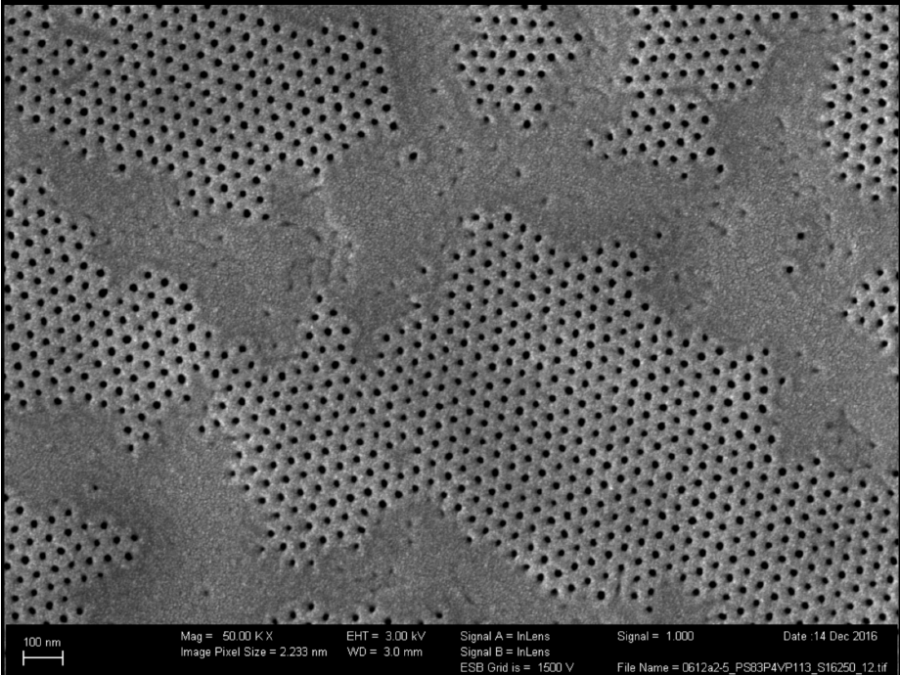
Connecting membrane pores and production parameters via machine learning (COMPUTING)
Isoporous block-copolymer membranes play a fundamental role in the filtration of liquids and can, for example, be used to purify drinking water. Despite recent progress in understanding the membrane formation process, finding suitable production parameters for a given precursor material (such as polymers of a certain length) still occurs in a trial-and-error fashion, wasting materials, […]